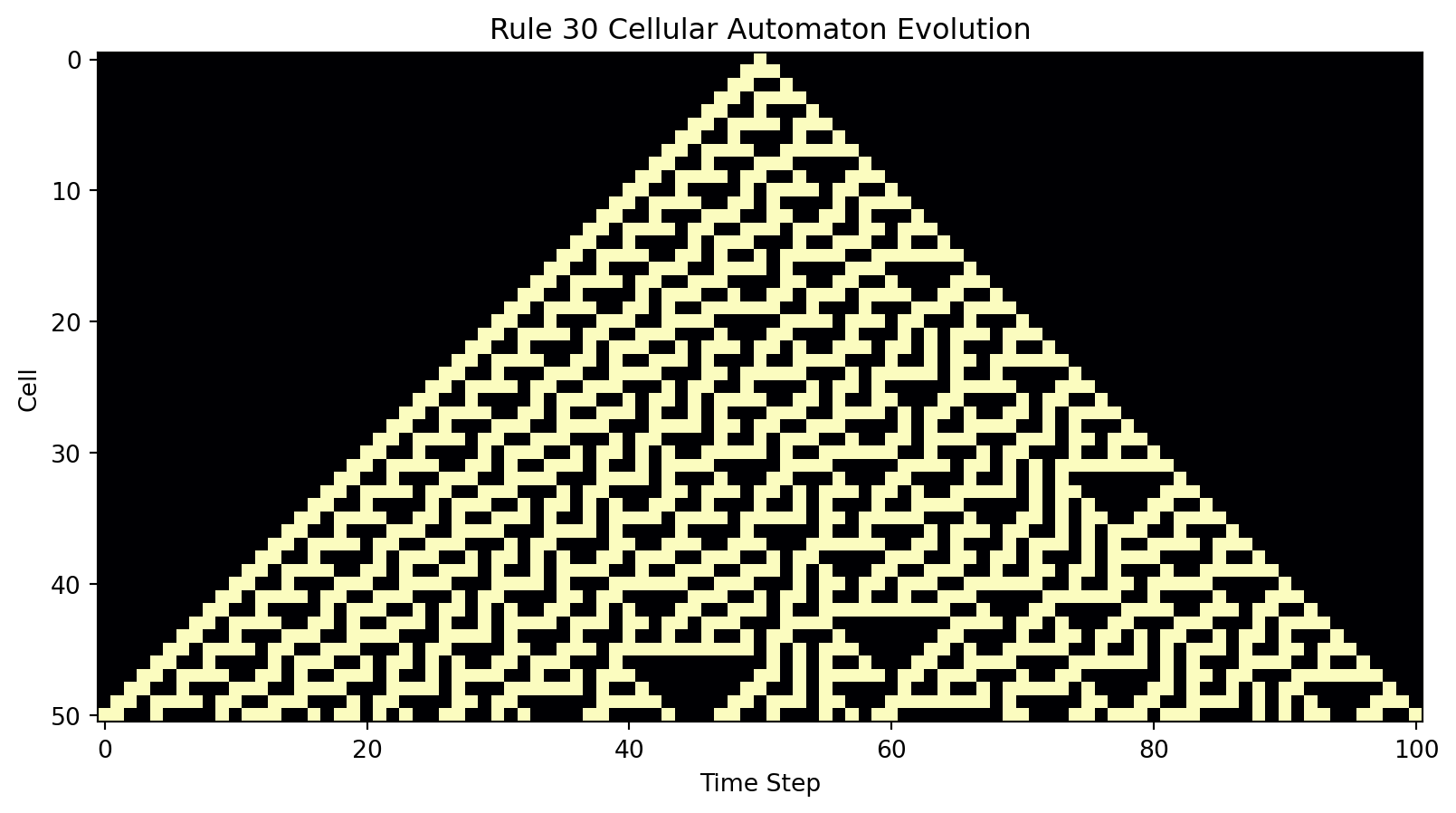
import numpy as np
import matplotlib.pyplot as plt
def apply_rule(rule, state, pos):
left = state[pos - 1]
center = state[pos]
right = state[(pos + 1) % len(state)]
out = rule[left * 4 + center * 2 + right]
return out
def evolve(rule, initial_state, steps):
state = initial_state.copy()
history = [state.copy()]
for _ in range(steps):
new_state = np.zeros_like(state)
for i in range(len(state)):
new_state[i] = apply_rule(rule, state, i)
state = new_state
history.append(state.copy())
return history
def plot_evolution(history):
plt.figure(figsize=(10, 5))
plt.imshow(history, cmap="magma", interpolation="nearest")
plt.xlabel("Time Step")
plt.ylabel("Cell")
plt.title("Rule 30 Cellular Automaton Evolution")
plt.show()
# Define Rule 30
rule30 = np.array([0, 1, 1, 1, 1, 0, 0, 0], dtype=int)
# Set initial state
initial_state = np.zeros(101, dtype=int)
initial_state[50] = 1
# Evolve and plot
steps = 50
evolution_history = evolve(rule30, initial_state, steps)
plot_evolution(np.array(evolution_history))